Cilkscale can help you reason about the parallel performance and scalability of
your Cilk program. Cilkscale enables you to:
- Collect statistics of parallel performance for your application.
- Measure the work, span, and parallelism of your (sub-)computations and predict how their performance will scale on
multiple processors.
- Automatically benchmark your program on different numbers of processors.
- Produce tables and graphical plots with the above performance and scalability
measurements.
This guide will walk you through the basic steps of profiling the parallel
performance and scalability of your Cilk application with Cilkscale. By the
end of this guide, you will know how to generate performance and scalability
tables and plots like the ones shown below and have a basic understanding of
how to use them to diagnose parallel performance limitations of your
application. For details on the Cilkscale components, user options, and output
information, see the Cilkscale reference page.
Note:
This guide assumes that OpenCilk is installed within
/opt/opencilk/ and that the OpenCilk C++ compiler can be invoked from the
terminal as /opt/opencilk/bin/clang++, as shown in this
example.
System setup for reported performance measurements:
All timings reported
in this page are measured on a laptop with an 8-core Intel Core i7-10875H CPU,
using OpenCilk 2.0.1 on Ubuntu 20.04 (via the Windows Subsystem for Linux v2 on
Windows 10).
Example application
We shall illustrate how to use the various components of Cilkscale with a
Cilk/C++ application that implements a parallel divide-and-conquer
quicksort. The source code for our
simple program, qsort.cpp, is shown below.
#include <iostream>
#include <algorithm>
#include <random>
#include <cilk/cilk.h>
constexpr std::ptrdiff_t BASE_CASE_LENGTH = 32;
template <typename T>
void sample_qsort(T* begin, T* end) {
if (end - begin < BASE_CASE_LENGTH) {
std::sort(begin, end);
} else {
--end;
T* middle = std::partition(begin, end,
[pivot=*end](T a) { return a < pivot; });
std::swap(*end, *middle);
cilk_scope {
cilk_spawn sample_qsort(begin, middle);
sample_qsort(++middle, ++end);
}
}
}
int main(int argc, char* argv[]) {
long n = 100 * 1000 * 1000;
if (argc == 2)
n = std::atoi(argv[1]);
std::default_random_engine rng;
std::uniform_int_distribution<long> dist(0,n);
std::vector<long> a(n);
std::generate(a.begin(), a.end(), [&]() { return dist(rng); });
std::cout << "Sorting " << n << " random integers" << std::endl;
sample_qsort(a.data(), a.data() + a.size());
bool pass = std::is_sorted(a.cbegin(), a.cend());
std::cout << "Sort " << ((pass) ? "succeeded" : "failed") << "\n";
return (pass) ? 0 : 1;
}
The qsort.cpp program simply generates a vector of pseudorandom numbers,
sorts it in parallel with the sample_qsort() function, and verifies the
result. We can compile and run it as follows.
$ /opt/opencilk/bin/clang++ qsort.cpp -fopencilk -O3 -o qsort
$ ./qsort 100000000
Sorting 100000000 random integers
Sort succeeded
Benchmarking and work/span analysis
Cilkscale instruments your Cilk program to collect performance measurements
during its execution. Cilkscale instrumentation operates in one of two modes:
- Benchmarking mode: Cilkscale measures the wall-clock execution time of your
program.
- Work/span analysis mode: Cilkscale measures the work, span, and parallelism of your program.
In either mode, you can use Cilkscale with two simple steps:
- Pass a Cilkscale instrumentation
flag
to the OpenCilk compiler when you compile and link your program. The result
is a Cilkscale-instrumented binary.
- Run the instrumented binary. Cilkscale collects performance measurements
and prints them to the standard output. (To output the report to a file,
set the
CILKSCALE_OUT
environment variable.) Your program otherwise runs as it normally would.
By default, Cilkscale only reports performance results for whole-program
execution. We will see how to perform fine-grained analyses of specific
sub-computations in the next section, after we show how to use Cilkscale in
benchmarking and work/span analysis mode.
Benchmarking instrumentation
To benchmark your application with Cilkscale, pass the
-fcilktool=cilkscale-benchmark flag to the OpenCilk compiler:
$ /opt/opencilk/bin/clang++ qsort.cpp -fopencilk -fcilktool=cilkscale-benchmark -O3 -o qsort_cs_bench
Running the instrumented binary now produces the program output as before,
followed by two lines with timing results in CSV
format:
$ ./qsort_cs_bench 100000000
[...]
tag,time (seconds)
,2.29345
The report table above contains a single, untagged row with the execution time
for the entire program. We will see shortly that if we use the Cilkscale API
for fine-grained analysis, then the report table will
contain additional rows.
Work/span analysis instrumentation
To analyze the parallel scalability of your application with Cilkscale, pass
the -fcilktool=cilkscale flag to the OpenCilk compiler:
$ /opt/opencilk/bin/clang++ qsort.cpp -fopencilk -fcilktool=cilkscale -O3 -o qsort_cs
When you run the instrumented binary, the program output is followed by the
Cilkscale work/span analysis report in CSV format:
$ ./qsort_cs 100000000
[...]
tag,work (seconds),span (seconds),parallelism,burdened_span (seconds),burdened_parallelism
,26.9397,2.29954,11.7153,2.29986,11.7136
The work, span, and parallelism measurements in the report depend on your
program's input and logical parallelism but not on the number of
processors on which it is run. The Cilkscale reference page describes the
specific quantities reported by
Cilkscale.
As before, the reported measurements above are untagged and refer to
whole-program execution.
Note:
The Cilkscale-instrumented binary in work/span analysis mode is
slower than its non-instrumented counterpart. The slowdown is generally no
larger than 10x and typically less than 2x. In the examples above, qsort and
qsort_cs_bench took 2.3s while qsort_cs took 3.4s (slowdown = 1.5x).
Fine-grained analysis
Cilkscale provides a C/C++
API for
benchmarking or analyzing specific regions in a program. The Cilkscale API
allows you to focus on and distinguish between specific parallel regions of
your computation when measuring its parallel performance and scalability.
Using the Cilkscale API is similar to using common C/C++ APIs for timing
regions of interest (such as the C++ std::chrono library or the POSIX
clock_gettime() function).
Let's see how we can use the Cilkscale API to analyze the execution of
sample_qsort() function in our example quicksort application. That is, we
want to exclude the computations for initializing a random vector of integers
or verifying the sort correctness, which are all executed serially anyway. To
achieve this, make the following three changes to the code.
-
Include the Cilkscale API header file. E.g., after line 4 in qsort.cpp:
#include <cilk/cilkscale.h>
-
Create work-span snapshots using calls to wsp_getworkspan() around the
region we want to analyze. E.g., around the call to sample_qsort() in
line 35 in qsort.cpp:
wsp_t start = wsp_getworkspan();
sample_qsort(a.data(), a.data() + a.size());
wsp_t end = wsp_getworkspan();
-
Evaluate the work and span between the relevant snapshots and print the
analysis results with a descriptive tag. E.g., just before the program
terminates in line 39 in qsort.cpp:
wsp_t elapsed = wsp_sub(end, start);
wsp_dump(elapsed, "qsort_sample");
Then, save the edited program (here, we save it as qsort_wsp.cpp), compile it
with Cilkscale instrumentation as before, and run it:
$ /opt/opencilk/bin/clang++ qsort_wsp.cpp -fopencilk -fcilktool=cilkscale -O3 -o qsort_wsp_cs
$ ./qsort_wsp_cs 100000000
[...]
tag,work (seconds),span (seconds),parallelism,burdened_span (seconds),burdened_parallelism
sample_qsort,26.1502,1.08122,24.1859,1.08153,24.1788
,27.3133,2.24433,12.1699,2.24465,12.1682
Notice that the Cilkscale report above now contains an additional row tagged
sample_qsort, which was output by the corresponding call to wsp_dump():
sample_qsort,26.1502,1.08122,24.1859,1.08153,24.1788
The last row in the Cilkscale report is always untagged and corresponds to
the execution of the whole program.
Note:
If you compile your code without a Cilkscale instrumentation flag,
calls to the Cilkscale API are effectively ignored with zero overhead.
For more detailed information on the Cilkscale C/C++ API, as well as an example
of how to aggregate work/span analysis measurements from disjoint code regions,
see the relevant section of the Cilkscale reference
page.
Automatic scalability benchmarks and visualization
Cilkscale includes a Python script which automates the process of benchmarking
and analyzing the scalability of your Cilk program. Specifically, the
Cilkscale Python script helps you do the following:
- Collect work/span analysis measurements for your program.
- Benchmark your program on different numbers of processors and collect
empirical scalability measurements.
- Store the combined analysis and benchmarking results in a CSV table.
- Visualize the analysis and benchmarking results with informative execution
time and speedup plots.
The Cilkscale Python script is found at share/Cilkscale_vis/cilkscale.py
within the OpenCilk installation directory.
Prerequisites:
To use the Cilkscale Python script, you need:
- Python 3.8 or later.
- (Optional) matplotlib 3.5.0 or later;
only required if producing graphical plots.
How to run
To use the cilkscale.py script, you must pass it two Cilkscale-instrumented
binaries of your program — one with -fcilktool=cilkscale-benchmark and one with
-fcilktool=cilkscale — along with a number of optional arguments.
For a description of the cilkscale.py script's arguments, see the Cilkscale reference page.
Let's now see an example of using the cilkscale.py script to analyze and
benchmark our qsort_wsp.cpp program, which uses the Cilkscale API to profile
the sample_qsort() function. First, we build the two Cilkscale-instrumented
binaries:
$ /opt/opencilk/bin/clang++ qsort_wsp.cpp -fopencilk -fcilktool=cilkscale-benchmark -O3 -o qsort_cs_bench
$ /opt/opencilk/bin/clang++ qsort_wsp.cpp -fopencilk -fcilktool=cilkscale -O3 -o qsort_cs
Then, we run cilkscale.py with our instrumented binaries on a sequence of
100,000,000 random integeres, and specify the output paths for the resulting
CSV table and PDF document of visualization plots:
$ python3 /opt/opencilk/share/Cilkscale_vis/cilkscale.py \
-c ./qsort_cs -b ./qsort_cs_bench \
-ocsv cstable_qsort.csv -oplot csplots_qsort.pdf \
--args 100000000
Terminal output
The cilkscale.py script first echoes the values for all of its parameters,
including unspecified parameters with default values:
Namespace(args=['100000000'], cilkscale='./qsort_cs', cilkscale_benchmark='./qsort_cs_bench', cpu_counts=None, output_csv='cstable_qsort.csv', output_plot='csplots_qsort.pdf', rows_to_plot='all')
Then, it runs the instrumented binary for work/span analysis on all available
cores and prints its standard output and standard error streams. You should
make sure that the program output is as expected.
>> STDOUT (./qsort_cs 100000000)
Sorting 100000000 random integers
Sort succeeded
<< END STDOUT
>> STDERR (./qsort_cs 100000000)
<< END STDERR
Once the work/span analysis pass is done, cilkscale.py runs the instrumented
binary for benchmarking on different numbers of processors. The number of
benchmarking runs and corresponding numbers of processors are determined by the
-cpus arguments to cilkcsale.py. (If this argument is not specified, the
program will run on processors, where is the number of
available physical cores in the system.)
INFO:runner:Generating scalability data for 8 cpus.
INFO:runner:CILK_NWORKERS=1 taskset -c 0 ./qsort_cs_bench 100000000
INFO:runner:CILK_NWORKERS=2 taskset -c 0,2 ./qsort_cs_bench 100000000
INFO:runner:CILK_NWORKERS=3 taskset -c 0,2,4 ./qsort_cs_bench 100000000
INFO:runner:CILK_NWORKERS=4 taskset -c 0,2,4,6 ./qsort_cs_bench 100000000
INFO:runner:CILK_NWORKERS=5 taskset -c 0,2,4,6,8 ./qsort_cs_bench 100000000
INFO:runner:CILK_NWORKERS=6 taskset -c 0,2,4,6,8,10 ./qsort_cs_bench 100000000
INFO:runner:CILK_NWORKERS=7 taskset -c 0,2,4,6,8,10,12 ./qsort_cs_bench 100000000
INFO:runner:CILK_NWORKERS=8 taskset -c 0,2,4,6,8,10,12,14 ./qsort_cs_bench 100000000
In this example, the program is benchmarked on up to 8 CPU cores with IDs 0, 2,
4, …. This is because cilkscale.py only uses distinct physical cores by
default. In the computer used for this example, core IDs 1, 3, 5, … correspond
to logical cores used in simultaneous
multithreading or
"hyper-threading".
Finally, cilkscale.py processes the collected benchmarking and work/span
analysis measurements and generates runtime and speedup plots for each analyzed
region (and the entire program).
INFO:plotter:Generating plot (2 subplots)
The Cilkscale benchmarking and scalability analysis reports are returned in
tabular and graphical form.
Tabular output
The raw measurements are output as a CSV table in the file pointed to by the
-ocsv argument to cilkscale.py. The CSV table contains, for each analyzed
region, the work/span analysis results and benchmark times for all numbers of
processors.
For example, the above run produced the following table:
$ cat cstable_qsort.csv
tag,work (seconds),span (seconds),parallelism,burdened_span (seconds),burdened_parallelism,1c time (seconds),2c time (seconds),3c time (seconds),4c time (seconds),5c time (seconds),6c time (seconds),7c time (seconds),8c time (seconds)
sample_qsort,26.5126,0.986602,26.8726,0.986927,26.8638,8.67705,4.6205,3.3648,2.75881,2.43091,2.1171,1.93193,1.7941
,27.6918,2.16583,12.7858,2.16616,12.7839,9.68071,5.52596,4.26341,3.65358,3.32762,3.02633,2.82155,2.67563
To see the table contents more clearly, you can import cstable_qsort.csv into
a spreadsheet (e.g., with LibreOffice) or
pretty-print it with command-line
tools:
$ cat cstable_qsort.csv | sed -e 's/^,/ ,/g' | column -s, -t | less -
1 tag work (seconds) span (seconds) parallelism burdened_span (seconds) burdened_parallelism 1c time (seconds) . . .
2 sample_qsort 26.5126 0.986602 26.8726 0.986927 26.8638 8.67705 . . .
3 27.6918 2.16583 12.7858 2.16616 12.7839 9.68071 . . .
Scalability plots
Cilkscale produces a set of scalability plots from the raw measurements in its
reported table. These plots are stored the PDF file pointed to by the -oplot
argument to cilkscale.py. Specifically, Cilkscale produces two figures for
each analyzed region (i.e., row in the CSV table): one which plots execution
time and one which plots parallel speedup. For a more detailed description of
these plots' contents, see the Cilkscale reference
page.
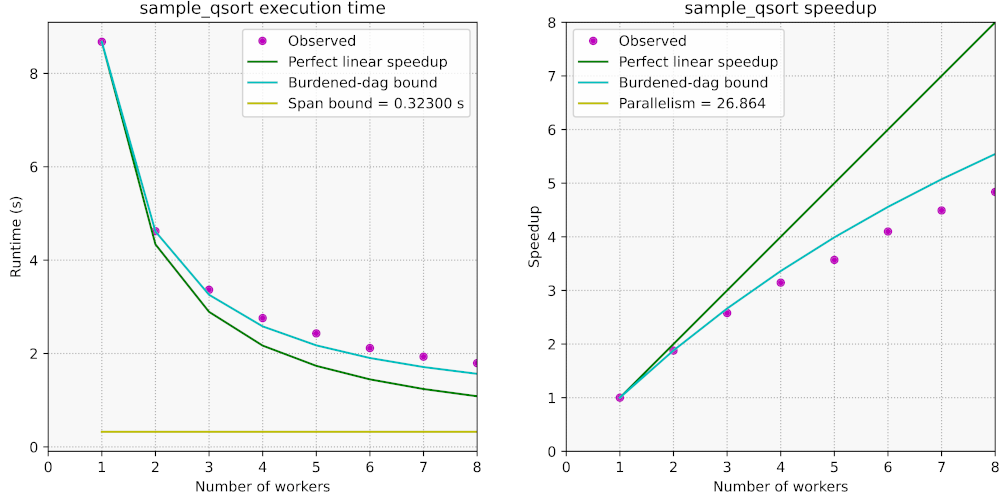
Here are the plots in csplots_qsort.pdf for the above example:
We have seen how to measure and explore the parallel performance and
scalability of a Cilk program. So... what next? How can we translate the
Cilkscale results into actionable insights on how to improve performance? As
with serial-program profiling, the answer varies somewhat depending on the
program at hand. We will return to this question with forthcoming
documentation and blog posts. Please let us know if
you'd like to be notified about important updates to OpenCilk and its
documentation.
In the meantime, we offer a brief discussion regarding the parallel scalability
of our qsort.cpp example, specifically the sample_qsort() function.
We observe the following:
- Our program shows sub-linear scalability. With 8 processor cores, the
parallel speedup is only about 4.9x.
- The observed performance roughly follows the burdened-dag bound and falls
short of it as the number of cores increases.
- The parallelism of
sample_qsort() is about 27, which is just over three
times as large as the amount of cores on the laptop where the experiments
were run.
A main issue with our parallel sample_qsort() is that it does not exhibit
sufficient parallelism. The parallelism of a computation upper-bounds the
number of processors that may productively work in parallel. Moreover,
computations with insufficient parallel slackness are typically
impacted adversely by scheduling and migration overheads. As a rule of thumb,
the parallelism of a computation is deemed sufficient if it is about 10x larger
(or more) than the number of available processors. On the other hand, if the
parallelism is too high — say, several orders of magnitude higher than the
number of processors — then the overhead for spawning tasks that are too
fine-grained may become substantial. In our case, the parallelism is low and
exhibits sufficient slackness for only 2–3 cores.
An additional issue is that the memory bandwidth of the laptop that was used in
these experiments becomes insufficient as more processing cores are used. This
is often the case for computations with low arithmetic intensity
when the observed parallel speedup falls below the burdened-dag speedup bound.
(Another possible cause for speedup below the burdened-dag bound is contention of parallel resources.) The memory bandwidth ceiling was
measured at about 24 GB/s with the
STREAM "copy" benchmark in either serial
or parallel mode.
If we want to improve the parallel performance of sample_qsort(), it appears
that our efforts, at least initially, are best spent increasing its
parallelism. One way to do that might be to undo the coarsening
of the base case (e.g., setting BASE_CASE_LENGTH = 1) but that makes the
recursion too fine-grained and introduces unnecessary spawning overhead — that
is, we may get better parallel speedup but slower execution overall. The one
remaining option then is to parallelize std::partition(), which is currently
serial and whose cost is linear with respect to the size of the input array.
We will not cover parallel partition algorithms for quicksort here, but warn
that designing and implementing efficient parallel partitions is an interesting
and nontrivial exercise!